K-Center From Scratch¶
This Jupyter Notebook provides a detailed, step-by-step guide on implementing the K-Center clustering algorithm from scratch. It covers the following key areas:
Sample data points
Select one initial cluster center
Update distance of each point to its closest center
Add a new cluster center
Show clutering results
[ ]:
import random
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
cmap = plt.get_cmap('tab10')
colors = [cmap(i) for i in range(cmap.N)]
mpl.rcParams["font.size"] = 24
mpl.rcParams["lines.linewidth"] = 2
seed = 0
random.seed(seed)
np.random.seed(seed)
n_samples = 10
markersize = 100
Sample data points¶
[ ]:
m1 = [1, 1]
cov1 = [[1, 0], [0, 1]]
s1 = np.random.multivariate_normal(m1, cov1, 5)
m2 = [-1, -1]
cov2 = [[1, -0.5], [-0.5, 1]]
s2 = np.random.multivariate_normal(m2, cov2, 5)
samples = np.concat([s1, s2])
np.random.shuffle(samples)
[ ]:
plt.figure(figsize=(5, 5))
plt.scatter(samples[:, 0], samples[:, 1], s=markersize, color="k")
# plt.axis("off")
plt.xlim([-2, 2])
plt.ylim([-2, 2])
plt.axis("equal")
(np.float64(-2.659691942232599),
np.float64(3.1307603678824707),
np.float64(-1.3757641112226862),
np.float64(3.4607340235073694))
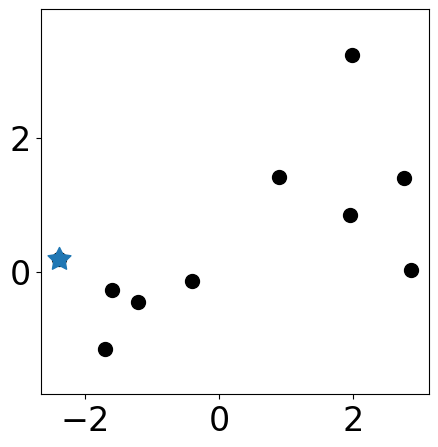
Select one initial cluster center¶
[ ]:
# initialize 1st center
centers = [0, ]
# initialize distance matrix
losses = []
n_points = samples.shape[0]
distances = np.full(n_points, np.inf)
[ ]:
plt.figure(figsize=(5, 5))
plt.scatter(samples[:, 0], samples[:, 1], s=markersize, color="k")
plt.scatter(samples[centers][:,0], samples[centers][:, 1],
s=markersize*3, c=colors[:len(centers)], marker=(5, 1))
plt.axis("equal")
(np.float64(-2.659691942232599),
np.float64(3.1307603678824707),
np.float64(-1.3757641112226862),
np.float64(3.4607340235073694))
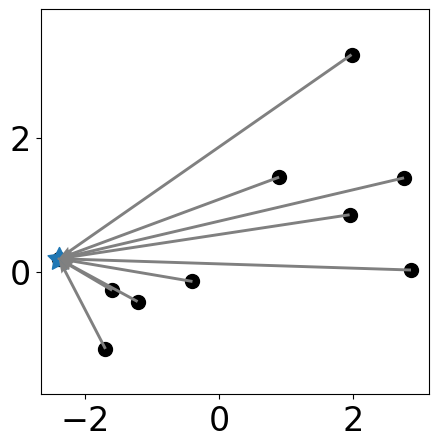
Update distance of each point to its closest center¶
[ ]:
last_center = centers[-1]
for i in range(n_points):
new_dist = np.linalg.norm(samples[i]-samples[last_center])
distances[i] = min(distances[i], new_dist)
Visualize Distances
[ ]:
plt.figure(figsize=(5, 5))
plt.scatter(samples[:, 0], samples[:, 1],
s=markersize, color="k")
# old centers
plt.scatter(samples[centers][:,0], samples[centers][:, 1], \
s=markersize*3, c=colors[:len(centers)], marker=(5, 1))
# assign predicted labels
labels = [np.argmin(np.sum((samples[i, :]-samples[centers])**2, axis=1)) \
for i in range(samples.shape[0])]
# add assignment vectors
for i in range(samples.shape[0]):
to_point = samples[centers[labels[i]]]
from_point = samples[i]
plt.quiver(from_point[0], from_point[1],
to_point[0]-from_point[0], to_point[1]-from_point[1],
angles="xy", scale_units="xy", scale=1, color="gray",
linewidth=1.5, linestyle="--")
plt.axis("equal")
(np.float64(-2.659691942232599),
np.float64(3.1307603678824707),
np.float64(-1.3757641112226862),
np.float64(3.4607340235073694))
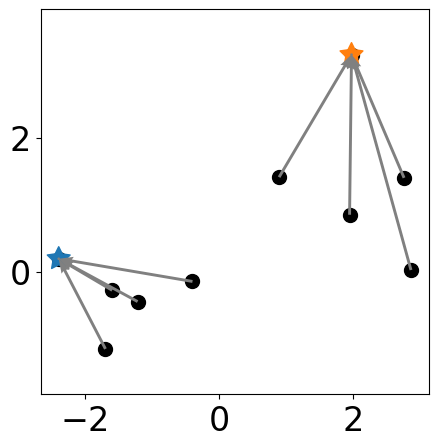
Add a new cluster center¶
[ ]:
## pick the point with the maximum distance as next center
next_center = np.argmax(distances)
centers.append(next_center)
[ ]:
plt.figure(figsize=(5, 5))
plt.scatter(samples[:, 0], samples[:, 1],
s=markersize, color="k")
# new centers
plt.scatter(samples[centers][:,0], samples[centers][:, 1], \
s=markersize*3, c=colors[:len(centers)], marker=(5, 1))
# assign predicted labels
labels = [np.argmin(np.sum((samples[i, :]-samples[centers])**2, axis=1)) \
for i in range(samples.shape[0])]
# add assignment vectors
for i in range(samples.shape[0]):
to_point = samples[centers[labels[i]]]
from_point = samples[i]
plt.quiver(from_point[0], from_point[1],
to_point[0]-from_point[0], to_point[1]-from_point[1],
angles="xy", scale_units="xy", scale=1, color="gray",
linewidth=1.5, linestyle="--")
plt.axis("equal")
(np.float64(-2.659691942232599),
np.float64(3.1307603678824707),
np.float64(-1.3757641112226862),
np.float64(3.4607340235073694))
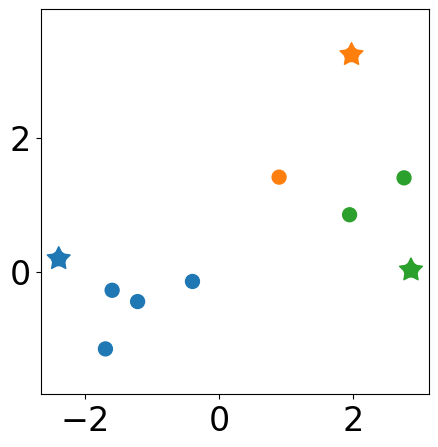
Show clutering results¶
[ ]:
def k_center(points, k):
losses = []
n_points = points.shape[0]
centers = []
distances = np.full(n_points, np.inf)
for j in range(0, k):
if j == 0:
centers = [0]
for i in range(n_points):
new_dist = np.linalg.norm(points[i]-points[centers[-1]])
distances[i] = min(distances[i], new_dist)
if j>=1:
next_center = np.argmax(distances)
centers.append(next_center)
losses.append(np.max(distances))
return centers, losses
[ ]:
k = 3
new_centers, losses = k_center(samples, k)
new_centers = samples[new_centers]
[ ]:
labels = [np.argmin(np.sum((samples[i, :]-new_centers)**2, axis=1)) for i in range(samples.shape[0])]
plt.figure(figsize=(5, 5))
plt.scatter(samples[:, 0], samples[:, 1],
s=markersize, color=[colors[l] for l in labels])
# new centers
plt.scatter(new_centers[:,0], new_centers[:, 1],
s=markersize*3, c=colors[:k], marker=(5, 1))
plt.axis("equal")
(np.float64(-2.659691942232599),
np.float64(3.1307603678824707),
np.float64(-1.3757641112226862),
np.float64(3.4607340235073694))
[ ]: