Predict Molecular Property using Recurrent Neural Networks¶
Install Dependencies¶
[ ]:
!pip install numpy pandas matplotlib seaborn torch torch_geometric rdkit wget tqdm
Download Dataset¶
[ ]:
!python -m \
wget https://raw.githubusercontent.com/deepchem/deepchem/master/datasets/delaney-processed.csv
Import Packages¶
[ ]:
import re
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib as mpl
import seaborn as sns
import tqdm
mpl.rcParams["font.size"] = 24
mpl.rcParams["lines.linewidth"] = 2
import rdkit.Chem as Chem
import torch
import torch.nn as nn
from torch.utils.data import Dataset, DataLoader
[ ]:
device = "cuda" if torch.cuda.is_available() else "cpu"
print("Device:", device)
Device: cuda
Load Dataset¶
[ ]:
DELANEY_FILE = "delaney-processed.csv"
TASK_COL = 'measured log solubility in mols per litre'
df = pd.read_csv(DELANEY_FILE)
print(f"Number of molecules in the dataset: {df.shape[0]}")
Number of molecules in the dataset: 1128
[ ]:
df["mol"] = df["smiles"].apply(lambda x: Chem.MolFromSmiles(x))
df = df[df["mol"].notna()]
[ ]:
# we will keep the following columns and discard the rest
df = df[["Compound ID", TASK_COL, "smiles", "mol"]]
df
| Compound ID | measured log solubility in mols per litre | smiles | mol | |
|---|---|---|---|---|
| 0 | Amigdalin | -0.770 | OCC3OC(OCC2OC(OC(C#N)c1ccccc1)C(O)C(O)C2O)C(O)... | <rdkit.Chem.rdchem.Mol object at 0x7f69d3dfc660> |
| 1 | Fenfuram | -3.300 | Cc1occc1C(=O)Nc2ccccc2 | <rdkit.Chem.rdchem.Mol object at 0x7f69d3dbd2a0> |
| 2 | citral | -2.060 | CC(C)=CCCC(C)=CC(=O) | <rdkit.Chem.rdchem.Mol object at 0x7f69d33e1cb0> |
| 3 | Picene | -7.870 | c1ccc2c(c1)ccc3c2ccc4c5ccccc5ccc43 | <rdkit.Chem.rdchem.Mol object at 0x7f69d33e1a80> |
| 4 | Thiophene | -1.330 | c1ccsc1 | <rdkit.Chem.rdchem.Mol object at 0x7f69d33a2570> |
| ... | ... | ... | ... | ... |
| 1123 | halothane | -1.710 | FC(F)(F)C(Cl)Br | <rdkit.Chem.rdchem.Mol object at 0x7f69d32f4350> |
| 1124 | Oxamyl | 0.106 | CNC(=O)ON=C(SC)C(=O)N(C)C | <rdkit.Chem.rdchem.Mol object at 0x7f69d32f43c0> |
| 1125 | Thiometon | -3.091 | CCSCCSP(=S)(OC)OC | <rdkit.Chem.rdchem.Mol object at 0x7f69d32f4430> |
| 1126 | 2-Methylbutane | -3.180 | CCC(C)C | <rdkit.Chem.rdchem.Mol object at 0x7f69d32f44a0> |
| 1127 | Stirofos | -4.522 | COP(=O)(OC)OC(=CCl)c1cc(Cl)c(Cl)cc1Cl | <rdkit.Chem.rdchem.Mol object at 0x7f69d32f4510> |
1128 rows × 4 columns
Create SMILES Dataset¶
In this notebook, we will use either rdkit CanonicalSMILES or augmented SMILES in training.
[ ]:
class SMILESDataset(Dataset):
def __init__(self, df, mol_col, target_col, augment=True):
self.all_df = df
self.mol_col = mol_col
self.target_col = target_col
self.augment = augment
def _featurize(self, mol):
#The mapping of the chemical compounds to SMILES strings are not unique
# "doRandom" allows the generation of different SMILES strings
# for the same chemical compounds.
# This is our data augmentation approach
smi = Chem.MolToSmiles(mol, doRandom=self.augment)
return smi
def __getitem__(self, idx):
mol = self.all_df.iloc[idx][self.mol_col]
smi = self._featurize(mol)
pka = self.all_df.iloc[idx][self.target_col]
return smi, float(pka)
def __len__(self):
return self.all_df.shape[0]
Padding to make all the SMILES strings have the same length
[ ]:
class Collate:
def __init__(self, tokenizer, pad_token="PAD"):
self.tokenizer = tokenizer
self.pad_token = pad_token
def collate(self, data):
smis = [smi for smi, pka in data]
X = [self.tokenizer.encode(smi) for smi in smis]
max_len = max([len(toks) for toks in X])
pad_idx = self.tokenizer.token2idx(self.pad_token)
# padded X
X = [[pad_idx]*(max_len-len(toks))+list(toks) for toks in X]
X = torch.Tensor(X).long()
Y = torch.Tensor([pka for smi, pka in data]).reshape(-1, 1).float()
return X, Y
Tokenizer converts a SMILE string to a list of integers. The integer \(i\) has the same meaning as Onehot Enbedding with the \(i\)-th position flagged as 1. We just convert to integers to save memory and be more compatible with nn.Embedding, which serves as a look-up table that converts the sparse Onehot embedding to a continuous embedding.
[ ]:
# convert characters to integers
class Tokenizer():
def __init__(self):
self.vocab = ["PAD", "UNK", "B", "Br", "Cl", "C", "H", \
"N", "O", "S", "P", "F", "I", \
"b", "c", "n", "o", "s", "p", "[", "]", \
"(", ")", ".", " ", "=", "#", \
"+", "-", ":", "~", "@", "*", "%", \
"/", "\\", "0", "1", "2", "3", "4", \
"5", "6", "7", "8", "9"]
self.i2v = {i: v for i, v in enumerate(self.vocab)}
self.v2i = {v:i for i, v in enumerate(self.vocab)}
SMI_REGEX_PATTERN = r"""(\[[^\]]+]|Br?|Cl?|N|O|S|P|F|I|b|c|n|o|s|p|\(|\)|\.|=|
#|-|\+|\\|\/|:|~|@|\?|>>?|\*|\$|\%[0-9]{2}|[0-9])"""
self.regex_pattern = SMI_REGEX_PATTERN
self.regex = re.compile(self.regex_pattern)
def token2idx(self, v):
if v in self.vocab:
return self.v2i[v]
else:
return self.v2i["UNK"]
def encode(self, smi):
lst = []
tokens = [token for token in self.regex.findall(smi)]
for v in tokens:
lst.append(self.token2idx(v))
return lst
Example of tokenizing “c1cccs1”
[ ]:
tokenizer = Tokenizer()
# smi = df.iloc[4]["smiles"]
smi = "c1cccs1"
# smi = "ClC(C)C"
print(smi)
tokens = [token for token in tokenizer.regex.findall(smi)]
print("Tokens:", tokens)
tok_ids = tokenizer.encode(smi)
print("Token ids:", tok_ids)
c1cccs1
Tokens: ['c', '1', 'c', 'c', 'c', 's', '1']
Token ids: [14, 37, 14, 14, 14, 17, 37]
Example of padding “c1cccs1”
[ ]:
Collate(tokenizer).collate([("c1cccs1", -1.33), ("ClC(C)C", -1.41)])
(tensor([[14, 37, 14, 14, 14, 17, 37],
[ 0, 4, 5, 21, 5, 22, 5]]),
tensor([[-1.3300],
[-1.4100]]))
Split Dataset¶
[ ]:
from sklearn.model_selection import train_test_split
# training/validation dataset
data_size = df.shape[0]
test_ratio = 0.10
test_size = int(data_size*test_ratio)
train_indices, test_indices = train_test_split(range(data_size), test_size=test_size, shuffle=True)
print(f"Training size: {len(train_indices)}, test size: {len(test_indices)}")
train_df, test_df = df.iloc[train_indices], df.iloc[test_indices]
Training size: 1016, test size: 112
[ ]:
# change to False in order to use rdkit CanonicalSMILES only
augment = True
batch_size = 32
tokenizer = Tokenizer()
print(tokenizer.vocab)
vocab_size = len(tokenizer.vocab)
print(f"Number of tokens: {vocab_size}")
## batching
instance = Collate(tokenizer)
collate_fn = instance.collate
train_data = SMILESDataset(train_df, mol_col="mol", target_col=TASK_COL, augment=augment)
train_loader = DataLoader(train_data, \
batch_size=batch_size, shuffle=True, collate_fn=collate_fn, drop_last=False)
test_data = SMILESDataset(test_df, mol_col="mol", target_col=TASK_COL, augment=augment)
test_loader = DataLoader(test_data, \
batch_size=batch_size, shuffle=False, collate_fn=collate_fn, drop_last=False)
['PAD', 'UNK', 'B', 'Br', 'Cl', 'C', 'H', 'N', 'O', 'S', 'P', 'F', 'I', 'b', 'c', 'n', 'o', 's', 'p', '[', ']', '(', ')', '.', ' ', '=', '#', '+', '-', ':', '~', '@', '*', '%', '/', '\\', '0', '1', '2', '3', '4', '5', '6', '7', '8', '9']
Number of tokens: 46
Example of calling the “train_data” function to generate a SMILES string and print its assocaited ground truth solubility.
[ ]:
train_data[0]
('c12ccccc2c(ccc1)N', -1.92)
calling the “train_data” function 5 times to generate 5 different SMILES strings for the same compound.
[ ]:
print([train_data[4] for i in range(5)])
[('CC(C(O)C)C', -0.18), ('CC(C)C(O)C', -0.18), ('CC(C(C)C)O', -0.18), ('CC(C(C)C)O', -0.18), ('OC(C)C(C)C', -0.18)]
We will print a small batch to see how the Dataloader pad for SMILES of different length. The following block shows that each batch contains two items: X of shape (batch_size, length) and Y of shape (batch_size, 1). length is the length of the longest SMILES in the batch. The other SMILES in the same batch will be padded to this length using 0 (the index of PAD in the Tokenizer). We will adopt pre-padding in this notebook, which means padding 0’s at the beginning.
[ ]:
batch = next(iter(DataLoader(train_data, \
batch_size=5, shuffle=True, collate_fn=collate_fn, drop_last=False)))
print(batch)
(tensor([[ 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 14, 37, 14, 21,
14, 21, 4, 22, 14, 14, 14, 37, 22, 7],
[ 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 14, 37, 14, 14, 21,
14, 14, 21, 14, 37, 3, 22, 3, 22, 3],
[ 5, 37, 38, 21, 11, 22, 5, 39, 21, 5, 22, 5, 25, 5, 5, 21, 5, 25,
5, 39, 5, 5, 5, 38, 5, 38, 5, 21, 5, 5, 37, 8, 22, 21, 5, 22,
5, 21, 5, 21, 5, 38, 22, 5, 22, 21, 8, 22, 5, 21, 25, 8, 22, 5,
8, 5, 21, 25, 8, 22, 5, 22, 25, 8],
[ 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 5, 21, 5, 22,
7, 37, 14, 38, 15, 14, 14, 14, 14, 38, 5, 21, 25, 9, 22, 7, 21, 5,
22, 14, 38, 14, 14, 14, 15, 14, 37, 38],
[ 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 15, 37, 21, 7, 22, 14, 21, 14, 21, 15, 15, 14, 37, 9, 5, 22, 5,
21, 5, 22, 21, 5, 22, 5, 22, 25, 8]]), tensor([[-1.5200],
[-4.5000],
[-4.9000],
[-4.6340],
[-2.2530]]))
Model¶
[ ]:
class SMILESModel(nn.Module):
def __init__(self, vocab_size, emb_dim, \
hidden_dim, n_layers, dropout=0.3, rnn_type="lstm", bidirectional=True):
super(SMILESModel, self).__init__()
#Convert each input token into an "emb_dim" vector.
self.embedding = nn.Embedding(vocab_size, emb_dim)
self.rnn_type = rnn_type
# choose rnn module
if self.rnn_type == "rnn":
self.rnn = nn.RNN(emb_dim, hidden_dim, n_layers, dropout=dropout,
batch_first=True, bidirectional=bidirectional)
elif self.rnn_type == "lstm":
self.rnn = nn.LSTM(emb_dim, hidden_dim, n_layers, dropout=dropout,
batch_first=True, bidirectional=bidirectional)
elif self.rnn_type == "gru":
self.rnn = nn.GRU(emb_dim, hidden_dim, n_layers, dropout=dropout,
batch_first=True, bidirectional=bidirectional)
else:
raise ValueError("rnn_type must be 'rnn', 'lstm', or 'gru'")
self.dropout = nn.Dropout(dropout)
if bidirectional:
self.fc = nn.Linear(2*hidden_dim, 1)
else:
self.fc = nn.Linear(hidden_dim, 1)
def forward(self, token_batch):
out = token_batch.long()
out = self.embedding(out) # convert ingeters to continuous embeddings
out, _ = self.rnn(out)
out = out[:, -1, :]
out = self.dropout(out)
out = self.fc(out)
return out
[ ]:
def train_one_epoch(model, criterion, optimizer, dataloader, clip):
model.train()
train_loss = []
for (x, y) in dataloader:
x, y = x.to(device), y.to(device)
optimizer.zero_grad()
y_pred = model(x)
loss = criterion(y_pred, y)
loss.backward()
nn.utils.clip_grad_norm_(model.parameters(), clip)
optimizer.step()
train_loss.append(loss.detach().cpu().numpy())
return np.mean(train_loss)
def val_one_epoch(model, criterion, dataloader):
model.eval()
val_loss = []
with torch.no_grad():
for (x, y) in dataloader:
x, y = x.to(device), y.to(device)
y_pred = model(x)
loss = criterion(y_pred, y)
val_loss.append(loss.detach().cpu().numpy())
return np.mean(val_loss)
Training¶
[ ]:
vocab_size = len(Tokenizer().vocab)
lr = 1e-3
emb_dim = 16
hidden_dim = 32
dropout = 0.3
n_layers = 4
clip = 5 # mitigate gradient exploding in BPTT
# bidirectional: run the sentence from "left to right" and from "right to left"
model = SMILESModel(vocab_size, emb_dim, hidden_dim,
n_layers, dropout=dropout, rnn_type="rnn", bidirectional=False)
model.to(device)
model = model.float()
print("Number of trainable parameters:",
sum(p.numel() for p in model.parameters() if p.requires_grad))
optimizer = torch.optim.Adam(model.parameters(), lr=lr)
criterion = nn.MSELoss()
n_epochs = 200
train_loss = []
val_loss = []
for epoch in tqdm.tqdm(range(n_epochs)):
losses = train_one_epoch(model, criterion, optimizer, train_loader, clip)
train_loss.append(np.mean(losses))
losses = val_one_epoch(model, criterion, test_loader)
val_loss.append(np.mean(losses))
Number of trainable parameters: 8705
100%|██████████| 200/200 [01:15<00:00, 2.66it/s]
[ ]:
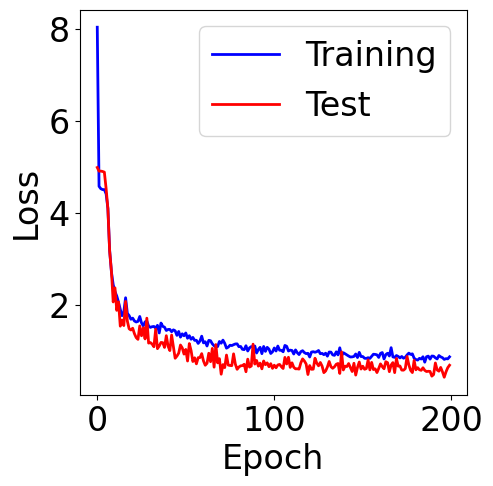
f, ax = plt.subplots(1, 1, figsize=(5,5))
ax.plot(train_loss, c="blue", label="Training")
ax.plot(val_loss, c="red", label="Test")
plt.xlabel("Epoch")
plt.ylabel("Loss")
plt.legend()
<matplotlib.legend.Legend at 0x7f69c8322210>
Evaluation Metrics¶
[ ]:
truths = []
predictions = []
model.eval()
with torch.no_grad():
for x, y in test_loader:
if device == "cuda":
x = x.to(device)
y_pred = model(x).reshape(-1)
predictions.extend(y_pred.cpu().detach().numpy().tolist())
y = y.reshape(y_pred.shape).cpu().numpy().tolist()
truths.extend(y)
[ ]:
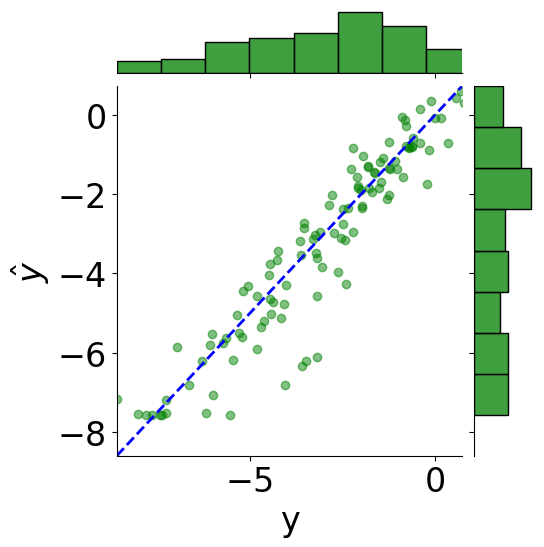
tmp_df = pd.DataFrame({"y": truths, r"$\hat{y}$": predictions})
# scatter plot
g = sns.JointGrid(x="y", y=r"$\hat{y}$", data=tmp_df)
g = g.plot_joint(plt.scatter, c="green", alpha=0.5)
# line: y_pred = y
y_line = np.linspace(np.min(truths), np.max(predictions), 200)
g.ax_joint.plot(y_line, y_line, color="blue", linestyle="--");
# histograms
g = g.plot_marginals(sns.histplot, data=df, color="green", kde=False)
g.ax_joint.set_xlim(np.min(y_line), np.max(y_line))
g.ax_joint.set_ylim(np.min(y_line), np.max(y_line))
plt.show()
[ ]:
from sklearn.metrics import r2_score
from sklearn.metrics import mean_squared_error
print(f"MSE: {mean_squared_error(truths, predictions):.2f}")
print(f"Coefficient of determination: {r2_score(truths, predictions):.2f}")
MSE: 0.68
Coefficient of determination: 0.87
[ ]: