[ ]:
!pip install wget
Techniques to Prevent Overfitting in Neural Networks¶
Dropout
Weight decay
Early stopping
[2]:
import numpy as np
import pandas as pd
import torch
import torch.nn as nn
import torch.nn.functional as F
import matplotlib as mpl
import matplotlib.pyplot as plt
from matplotlib.colors import ListedColormap
from sklearn.metrics import confusion_matrix
from sklearn.metrics import RocCurveDisplay, auc
from sklearn.model_selection import train_test_split
cmap = plt.get_cmap('tab10')
colors = [cmap(i) for i in range(cmap.N)]
mpl.rcParams["font.size"] = 24
mpl.rcParams["lines.linewidth"] = 2
Load Dataset¶
[ ]:
!python -m wget https://raw.githubusercontent.com/deepchem/deepchem/master/datasets/delaney-processed.csv \
--output delaney-processed.csv
[4]:
DELANEY_FILE = "delaney-processed.csv"
TASK_COL = 'measured log solubility in mols per litre'
df = pd.read_csv(DELANEY_FILE)
print(f"Number of molecules in the dataset: {df.shape[0]}")
Number of molecules in the dataset: 1128
[5]:
X = df[[
"Molecular Weight",
"Number of H-Bond Donors",
"Number of Rings",
"Polar Surface Area"]].values
Y = df[TASK_COL].values.reshape(-1, 1) >= -2
print("Shape of X:", X.shape)
print("Shape of Y:", Y.shape)
Shape of X: (1128, 4)
Shape of Y: (1128, 1)
[6]:
device = "cuda" if torch.cuda.is_available() else "cpu"
print("Device:", device)
Device: cpu
MLP Model¶
[7]:
class MLPRegresion(torch.nn.Module):
def __init__(self, in_dim, hidden_dim, out_dim, norm=False, dropout=None):
super(MLPRegresion, self).__init__()
hidden_dim = [in_dim] + hidden_dim
nn = []
if norm:
nn.append(torch.nn.BatchNorm1d(in_dim))
for i in range(1, len(hidden_dim)):
nn.append(torch.nn.Linear(hidden_dim[i-1], hidden_dim[i]))
if dropout is not None:
nn.append(torch.nn.Dropout(dropout))
nn.append(torch.nn.ReLU())
nn.append(torch.nn.Linear(hidden_dim[-1], out_dim))
self.model = torch.nn.Sequential(*nn)
def forward( self, x):
for layer in self.model:
x = layer(x)
return x
Training Utils¶
[8]:
def train_one_epcoh(model, criterion, optimizer, dataloader):
losses = []
model.train()
for x, y_true in dataloader:
if device == "cuda":
x, y_true = x.to(device), y_true.to(device)
x, y_true = x.float(), y_true.float()
optimizer.zero_grad()
y_pred = model(x)
y_pred = torch.sigmoid(y_pred)
loss = criterion(y_pred, y_true.reshape(y_pred.shape))
loss.backward()
optimizer.step()
losses.append(loss.cpu().detach().item())
return losses
def val_one_epcoh(model, criterion, dataloader):
losses = []
model.eval()
with torch.no_grad():
for x, y_true in dataloader:
if device == "cuda":
x, y_true = x.to(device), y_true.to(device)
x, y_true = x.float(), y_true.float()
y_pred = model(x)
y_pred = torch.sigmoid(y_pred)
loss = criterion(y_pred, y_true.reshape(y_pred.shape))
losses.append(loss.cpu().detach().item())
return losses
[9]:
def sample_dataloaders(batch_size=128):
# training/validation dataset
test_size = int(len(X)*0.3)
X_train, X_test, y_train, y_test = train_test_split(X, Y, test_size=test_size, shuffle=True)
# create dataloaders
X_train, X_test, y_train, y_test = map(torch.tensor, (X_train, X_test, y_train, y_test))
train_data = torch.utils.data.TensorDataset(X_train, y_train)
train_loader = torch.utils.data.DataLoader(train_data, batch_size=batch_size,
shuffle=True, drop_last=False)
test_data = torch.utils.data.TensorDataset(X_test, y_test)
test_loader = torch.utils.data.DataLoader(test_data, batch_size=batch_size,
shuffle=False, drop_last=False)
return train_loader, test_loader
[10]:
def run_one_fold(model, criterion, optimizer, train_loader, test_loader, n_epochs, early_stop=None):
train_loss = []
val_loss = []
for epoch in range(n_epochs):
losses = train_one_epcoh(model, criterion, optimizer, train_loader)
train_loss.append(np.mean(losses))
losses = val_one_epcoh(model, criterion, test_loader)
val_loss.append(np.mean(losses))
if early_stop is not None:
early_stop(np.mean(losses), model)
if early_stop.early_stop:
print("Early stopping")
break
return train_loss, val_loss
[11]:
def evaluate(model, test_loader):
model.eval()
truth = []
preds = []
with torch.no_grad():
for x, y_true in test_loader:
if device == "cuda":
x, y_true = x.to(device), y_true.to(device)
x, y_true = x.float(), y_true.float()
truth.extend(y_true.cpu().detach().numpy().tolist())
output = model(x)
y_pred = torch.sigmoid(output).reshape(-1)
y_pred = y_pred.cpu().detach().numpy()
preds.extend(y_pred.tolist())
return truth, preds
Binary Classification Model to test teachniques to avoid overfitting¶
Dropout, Ridge Regularization (weight decay)¶
[12]:
n_splits = 3 # K-fold cross validation
n_epochs = 1500
batchzise = 32
hidden_dims = [512, 256, 256, 128, 128]
lr = 0.1
norm = True
## Feel free to tune hyperparameters below
# weight_decay = 1e-3
weight_decay = 0
# dropout = None
dropout = 0.5
Run cross validation.
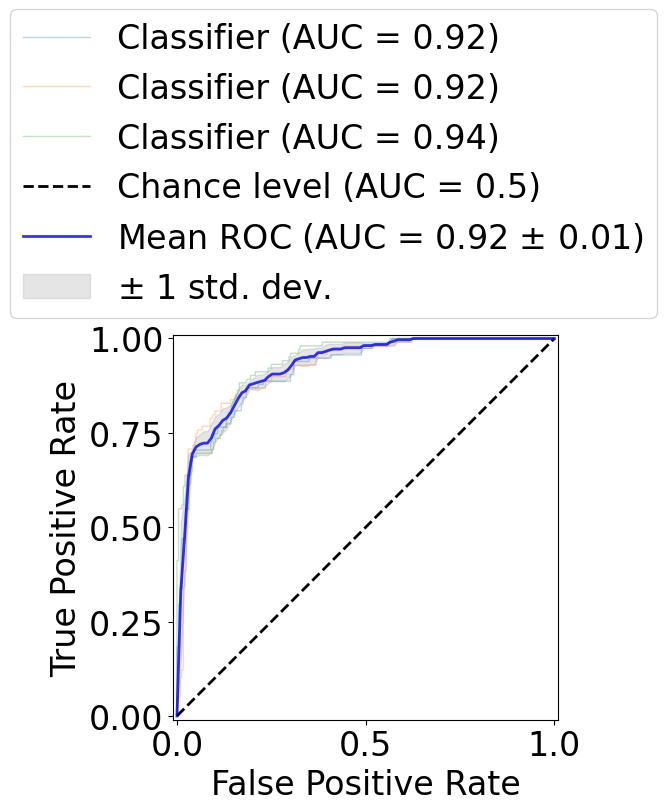
[13]:
fig, ax = plt.subplots(1, 1, figsize=(5, 5))
tprs = []
aucs = []
mean_fpr = np.linspace(0, 1, 100)
train_list = []
test_list = []
for fold in range(n_splits):
# define model
model = MLPRegresion(X.shape[1], hidden_dims, Y.shape[1], norm, dropout)
model = model.to(device)
model = model.float()
criterion = torch.nn.BCELoss()
optimizer = torch.optim.SGD(model.parameters(), lr=lr, weight_decay=weight_decay)
# train
train_loader, test_loader = sample_dataloaders(batchzise)
train_loss, val_loss = run_one_fold(model, criterion, optimizer,
train_loader, test_loader, n_epochs)
train_list.append(train_loss)
test_list.append(val_loss)
# evaluate
y_test_true, y_test_pred = evaluate(model, test_loader)
# record ROC curve
viz = RocCurveDisplay.from_predictions(
np.array(y_test_true),
np.array(y_test_pred),
alpha=0.3,
lw=1,
ax=ax,
plot_chance_level=(fold == n_splits-1),
)
interp_tpr = np.interp(mean_fpr, viz.fpr, viz.tpr)
interp_tpr[0] = 0.0
tprs.append(interp_tpr)
aucs.append(viz.roc_auc)
# plot
mean_tpr = np.mean(tprs, axis=0)
mean_tpr[-1] = 1.0
mean_auc = auc(mean_fpr, mean_tpr)
std_auc = np.std(aucs)
ax.plot(
mean_fpr,
mean_tpr,
color="b",
label=r"Mean ROC (AUC = %0.2f $\pm$ %0.2f)" % (mean_auc, std_auc),
lw=2,
alpha=0.8,
)
std_tpr = np.std(tprs, axis=0)
tprs_upper = np.minimum(mean_tpr + std_tpr, 1)
tprs_lower = np.maximum(mean_tpr - std_tpr, 0)
ax.fill_between(
mean_fpr,
tprs_lower,
tprs_upper,
color="grey",
alpha=0.2,
label=r"$\pm$ 1 std. dev.",
)
ax.set(
xlabel="False Positive Rate",
ylabel="True Positive Rate",
)
ax.legend(loc="lower right", bbox_to_anchor=(1.3, 1))
plt.show()
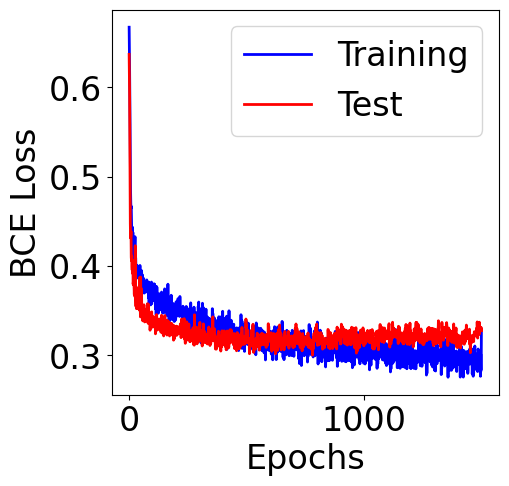
[14]:
plt.figure(figsize=(5, 5))
train_mean = np.vstack([np.array(l).reshape(1, -1) for l in train_list]).mean(axis=0)
train_std = np.vstack([np.array(l).reshape(1, -1) for l in train_list]).std(axis=0)
plt.plot(range(n_epochs), train_mean, color="b", label="Training")
# plt.fill_between(range(n_epochs),
# train_mean-train_std, train_mean+train_std,
# alpha=0.5, color="b")
test_mean = np.vstack([np.array(l).reshape(1, -1) for l in test_list]).mean(axis=0)
test_std = np.vstack([np.array(l).reshape(1, -1) for l in test_list]).std(axis=0)
plt.plot(range(n_epochs), test_mean, color="red", label="Test")
# plt.fill_between(range(n_epochs),
# test_mean-test_std, test_mean+test_std,
# alpha=0.5, color="r")
plt.xlabel("Epochs")
plt.ylabel("BCE Loss")
plt.legend()
[14]:
<matplotlib.legend.Legend at 0x78e1d7693fd0>
Early Stopping¶
[15]:
lr = 0.1
weight_decay = 0
dropout = None
norm = True
[16]:
class EarlyStopping:
"""Early stops the training if validation loss doesn't improve after a given patience."""
def __init__(self, patience=3, delta=1e-3, verbose=0):
"""
Params
------
patience : int
steps/ epochs to wait after last time validation loss improved
delta : float
minimum change in the monitored quantity to qualify as an improvement
path : str
path to checkpoint params
"""
self.patience = patience
self.counter = 0
self.best_score = None
self.early_stop = False
self.val_loss_min = np.inf
self.delta = delta
self.verbose = verbose
self.scores = []
def __call__(self, val_loss, model):
score = -val_loss
self.scores.append(score)
if self.best_score is None:
self.best_score = score
elif score < self.best_score + self.delta:
self.counter += 1
if self.verbose > 0:
print(f'EarlyStopping counter: {self.counter} out of {self.patience}')
if self.counter >= self.patience:
self.early_stop = True
else:
self.best_score = score
self.counter = 0
[17]:
patience = 30 # Early Stopping if validation loss does not increase for 30 epochs
delta = 1e-5
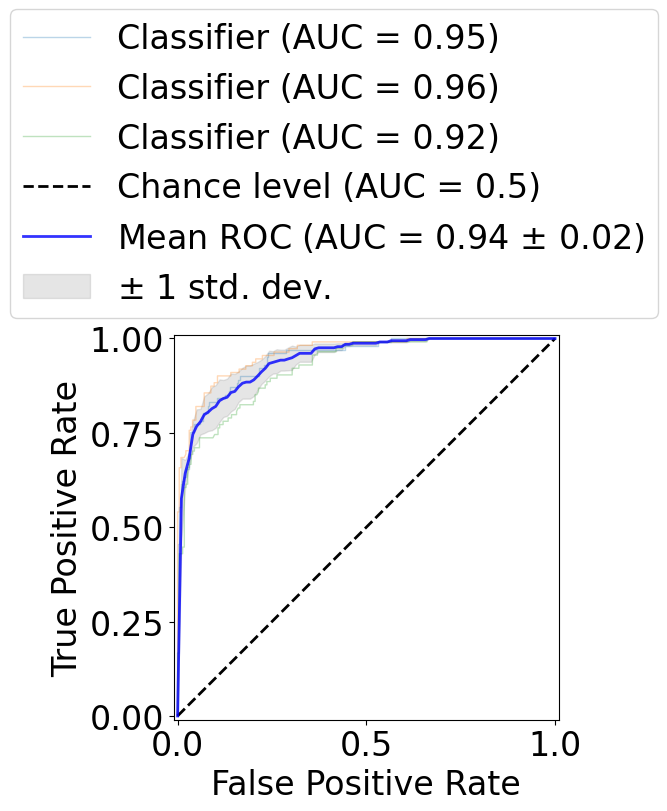
Run cross validation.
[18]:
fig, ax = plt.subplots(1, 1, figsize=(5, 5))
tprs = []
aucs = []
mean_fpr = np.linspace(0, 1, 100)
train_list = []
test_list = []
for fold in range(n_splits):
early_stopping = EarlyStopping(patience=patience, delta=delta)
# define model
model = MLPRegresion(X.shape[1], hidden_dims, Y.shape[1], norm, dropout)
model = model.to(device)
model = model.float()
criterion = torch.nn.BCELoss()
optimizer = torch.optim.SGD(model.parameters(), lr=lr, weight_decay=weight_decay)
# train
train_loader, test_loader = sample_dataloaders(batchzise)
train_loss, val_loss = run_one_fold(model, criterion, optimizer,
train_loader, test_loader, n_epochs, early_stopping)
train_list.append(train_loss)
test_list.append(val_loss)
# evaluate
y_test_true, y_test_pred = evaluate(model, test_loader)
# record ROC curve
viz = RocCurveDisplay.from_predictions(
np.array(y_test_true),
np.array(y_test_pred),
alpha=0.3,
lw=1,
ax=ax,
plot_chance_level=(fold == n_splits-1),
)
interp_tpr = np.interp(mean_fpr, viz.fpr, viz.tpr)
interp_tpr[0] = 0.0
tprs.append(interp_tpr)
aucs.append(viz.roc_auc)
# plot
mean_tpr = np.mean(tprs, axis=0)
mean_tpr[-1] = 1.0
mean_auc = auc(mean_fpr, mean_tpr)
std_auc = np.std(aucs)
ax.plot(
mean_fpr,
mean_tpr,
color="b",
label=r"Mean ROC (AUC = %0.2f $\pm$ %0.2f)" % (mean_auc, std_auc),
lw=2,
alpha=0.8,
)
std_tpr = np.std(tprs, axis=0)
tprs_upper = np.minimum(mean_tpr + std_tpr, 1)
tprs_lower = np.maximum(mean_tpr - std_tpr, 0)
ax.fill_between(
mean_fpr,
tprs_lower,
tprs_upper,
color="grey",
alpha=0.2,
label=r"$\pm$ 1 std. dev.",
)
ax.set(
xlabel="False Positive Rate",
ylabel="True Positive Rate",
)
ax.legend(loc="lower right", bbox_to_anchor=(1.3, 1))
plt.show()
Early stopping
Early stopping
Early stopping
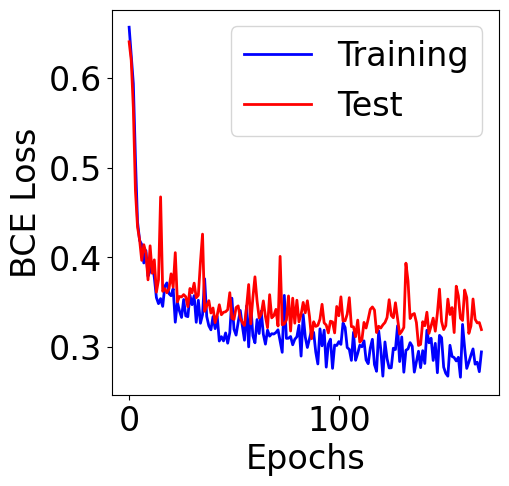
[19]:
plt.figure(figsize=(5, 5))
plt.plot(train_loss, color="b", label="Training")
plt.plot(val_loss, color="red", label="Test")
plt.xlabel("Epochs")
plt.ylabel("BCE Loss")
plt.legend()
[19]:
<matplotlib.legend.Legend at 0x78e1d3c43fd0>
[19]: