[ ]:
!pip install wget
K-nearest Neighbour¶
[2]:
import pandas as pd
import numpy as np
from matplotlib import pyplot as plt
Download Datasets¶
[3]:
!python -m wget https://raw.githubusercontent.com/xuhuihuang/uwmadisonchem361/refs/heads/main/delaney_dataset_200compounds.csv \
--output delaney_dataset_200compounds.csv
!python -m wget https://raw.githubusercontent.com/xuhuihuang/uwmadisonchem361/refs/heads/main/delaney_dataset_40compounds.csv \
--output delaney_dataset_40compounds.csv
!python -m wget https://raw.githubusercontent.com/xuhuihuang/uwmadisonchem361/refs/heads/main/delaney_dataset_44compounds_with_outliers.csv \
--output delaney_dataset_44compounds_with_outliers.csv
Saved under delaney_dataset_200compounds.csv
Saved under delaney_dataset_40compounds.csv
Saved under delaney_dataset_44compounds_with_outliers.csv
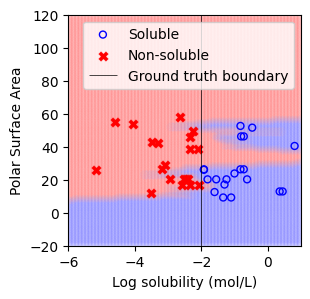
Load the curated Delaney dataset, which contains 40 compounds:¶
20 Soluble Compounds: Defined as those with a “measured log solubility in mols per litre” ≥ -2, labeled as 1.
20 Non-Soluble Compounds: Defined as those with a “measured log solubility in mols per litre” < -2, labeled as -1.
[4]:
df = pd.read_csv('delaney_dataset_40compounds.csv')
df.head(2)
[4]:
| Molecular Weight | Polar Surface Area | measured log solubility in mols per litre | solubility labels | smiles | |
|---|---|---|---|---|---|
| 0 | 103.124 | 23.79 | -1.00 | 1 | N#Cc1ccccc1 |
| 1 | 116.204 | 20.23 | -1.81 | 1 | CCCCCCCO |
[5]:
data = df.iloc[:].values
[6]:
# data with log solubility and Polar Surface Are as features.
X = data[:,[2,1]]
# solubility labels
y = data[:,3].astype(int)
[7]:
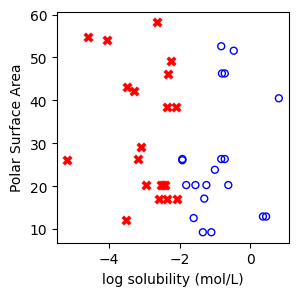
f, ax = plt.subplots(1,1,figsize=(3,3))
ax.scatter(X[np.where(y==1)[0],0],X[np.where(y==1)[0],1],s=25, marker='o', facecolors='none', edgecolor="blue", label='soluble')
ax.scatter(X[np.where(y==-1)[0],0],X[np.where(y==-1)[0],1],s=50, marker='X', color='red',linewidths=0.1, label='non-soluble')
ax.set_xlabel("log solubility (mol/L)")
ax.set_ylabel("Polar Surface Area")
#plt.legend()
[7]:
Text(0, 0.5, 'Polar Surface Area')
Let’s fit a 1-NN model¶
[8]:
from sklearn.neighbors import KNeighborsClassifier
neigh = KNeighborsClassifier(n_neighbors=1)
neigh.fit(X, y)
[8]:
KNeighborsClassifier(n_neighbors=1)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
KNeighborsClassifier(n_neighbors=1)