[ ]:
!pip install wget
Logistic regression¶
[2]:
import pandas as pd
import numpy as np
from matplotlib import pyplot as plt
Download Datasets¶
[ ]:
!python -m wget https://raw.githubusercontent.com/xuhuihuang/uwmadisonchem361/refs/heads/main/delaney_dataset_200compounds.csv \
--output delaney_dataset_200compounds.csv
!python -m wget https://raw.githubusercontent.com/xuhuihuang/uwmadisonchem361/refs/heads/main/delaney_dataset_40compounds.csv \
--output delaney_dataset_40compounds.csv
!python -m wget https://raw.githubusercontent.com/xuhuihuang/uwmadisonchem361/refs/heads/main/delaney_dataset_44compounds_with_outliers.csv \
--output delaney_dataset_44compounds_with_outliers.csv
Load the curated Delaney dataset, which contains 40 compounds:¶
20 Soluble Compounds: Defined as those with a “measured log solubility in mols per litre” ≥ -2, labeled as 1.
20 Non-Soluble Compounds: Defined as those with a “measured log solubility in mols per litre” < -2, labeled as -1.
[4]:
df = pd.read_csv('delaney_dataset_40compounds.csv')
df.head(2)
[4]:
| Molecular Weight | Polar Surface Area | measured log solubility in mols per litre | solubility labels | smiles | |
|---|---|---|---|---|---|
| 0 | 103.124 | 23.79 | -1.00 | 1 | N#Cc1ccccc1 |
| 1 | 116.204 | 20.23 | -1.81 | 1 | CCCCCCCO |
[5]:
data = df.iloc[:].values
[6]:
# data with Molecular Weight and Polar Surface Are as features.
X = data[:,0:2]
# solubility labels
y = data[:,3].astype(int)
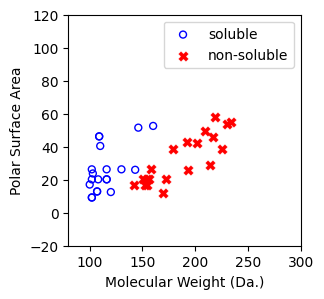
visualize the 40 compounds¶
[7]:
f, ax = plt.subplots(1,1,figsize=(3,3))
ax.scatter(X[np.where(y==1)[0],0],X[np.where(y==1)[0],1],s=25, marker='o', facecolors='none', edgecolor="blue", label='soluble')
ax.scatter(X[np.where(y==-1)[0],0],X[np.where(y==-1)[0],1],s=50, marker='X', color='red',linewidths=0.1, label='non-soluble')
ax.set_xlabel("Molecular Weight (Da.)")
ax.set_ylabel("Polar Surface Area")
ax.set_xlim(80,300)
ax.set_ylim(-20,120)
plt.legend()
[7]:
<matplotlib.legend.Legend at 0x7cbbfe13aa80>
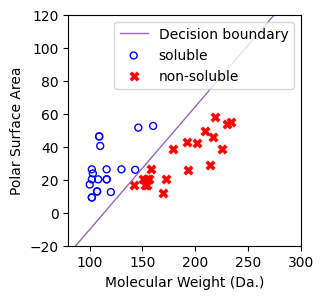
Fit a logistic regression model using sklearn¶
[8]:
# plot the decision boundary
def predict_boundary(x,regressor):
y = [(- regressor.coef_[0][0]*x[i] - regressor.intercept_)/(regressor.coef_[0][1]) for i in range(len(x))]
return y
# coef_: Coefficient of the features in the decision function.
# coef_: Shape of (1, n_features) for binary classification - \theta_1 and \theta_2
# intercept_: \thetha_0
# Decision boundary line with x in the range of [80, 300].
# Decison line: p=0, so that y (polar surface) = [(-\theta_1*x (molecualr weight) - \theta_0)/\theta_1
[9]:
from sklearn.linear_model import LogisticRegression
regressor = LogisticRegression(random_state=0,penalty=None).fit(X, y)
[10]:
f, ax = plt.subplots(1,1,figsize=(3,3))
ax.plot(
[80,300],
predict_boundary([80,300],regressor=regressor),
linewidth=1,
color="tab:purple",
label="Decision boundary",
)
ax.scatter(X[np.where(y==1)[0],0],X[np.where(y==1)[0],1],s=25, marker='o', facecolors='none', edgecolor="blue", label='soluble')
ax.scatter(X[np.where(y==-1)[0],0],X[np.where(y==-1)[0],1],s=50, marker='X', color='red',linewidths=0.1, label='non-soluble')
ax.set_xlabel("Molecular Weight (Da.)")
ax.set_ylabel("Polar Surface Area")
ax.set_xlim(80,300)
ax.set_ylim(-20,120)
plt.legend()
[10]:
<matplotlib.legend.Legend at 0x7cbbf1574770>
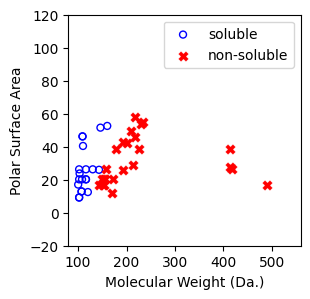
Logistic regression is robust to outliers¶
[11]:
df = pd.read_csv('delaney_dataset_44compounds_with_outliers.csv')
df.head(2)
data = df.iloc[:].values
# data with Molecular Weight and Polar Surface Are as features.
X = data[:,0:2]
# solubility labels
y = data[:,3].astype(int)
[12]:
f, ax = plt.subplots(1,1,figsize=(3,3))
ax.scatter(X[np.where(y==1)[0],0],X[np.where(y==1)[0],1],s=25, marker='o', facecolors='none', edgecolor="blue", label='soluble')
ax.scatter(X[np.where(y==-1)[0],0],X[np.where(y==-1)[0],1],s=50, marker='X', color='red',linewidths=0.1, label='non-soluble')
ax.set_xlabel("Molecular Weight (Da.)")
ax.set_ylabel("Polar Surface Area")
ax.set_xlim(80,560)
ax.set_ylim(-20,120)
plt.legend()
[12]:
<matplotlib.legend.Legend at 0x7cbbf0ec40e0>
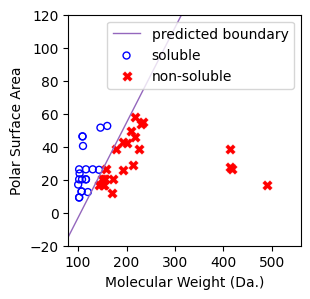
[13]:
from sklearn.linear_model import LogisticRegression
regressor = LogisticRegression(random_state=0,penalty=None).fit(X, y)
[14]:
f, ax = plt.subplots(1,1,figsize=(3,3))
ax.plot(
[80,560],
predict_boundary([80,560],regressor=regressor),
linewidth=1,
color="tab:purple",
label="predicted boundary",
)
ax.scatter(X[np.where(y==1)[0],0],X[np.where(y==1)[0],1],s=25, marker='o', facecolors='none', edgecolor="blue", label='soluble')
ax.scatter(X[np.where(y==-1)[0],0],X[np.where(y==-1)[0],1],s=50, marker='X', color='red',linewidths=0.1, label='non-soluble')
ax.set_xlabel("Molecular Weight (Da.)")
ax.set_ylabel("Polar Surface Area")
ax.set_xlim(80,560)
ax.set_ylim(-20,120)
plt.legend()
[14]:
<matplotlib.legend.Legend at 0x7cbbf1440c20>