Predict Molecular Property using Graph Neural Network¶
Install Dependencies¶
[ ]:
!pip install seaborn torch torch_geometric rdkit wget tqdm
Download Dataset¶
[ ]:
!python -m \
wget https://raw.githubusercontent.com/deepchem/deepchem/master/datasets/delaney-processed.csv
Saved under delaney-processed.csv
Import Packages¶
[ ]:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib as mpl
import seaborn as sns
import matplotlib.patches as patches
mpl.rcParams["font.size"] = 24
mpl.rcParams["lines.linewidth"] = 2
import tqdm
import rdkit.Chem as Chem
import rdkit.Chem.AllChem as AllChem
import torch
import torch.nn as nn
from torch_geometric.data import Data
from torch_geometric.data import Dataset
from torch_geometric.loader import DataLoader
from torch_geometric.nn.conv import GCNConv
from torch_geometric.nn.pool import global_mean_pool
[ ]:
device = "cuda" if torch.cuda.is_available() else "cpu"
print("Device:", device)
Device: cuda
Load Dataset¶
[ ]:
DELANEY_FILE = "delaney-processed.csv"
TASK_COL = 'measured log solubility in mols per litre'
df = pd.read_csv(DELANEY_FILE)
print(f"Number of molecules in the dataset: {df.shape[0]}")
Number of molecules in the dataset: 1128
Create molecule from SMILES
[ ]:
df["mol"] = df["smiles"].apply(lambda x: Chem.MolFromSmiles(x))
df = df[df["mol"].notna()]
[ ]:
# we will keep the following columns and discard the rest
df = df[["Compound ID", TASK_COL, "smiles", "mol"]]
df
| Compound ID | measured log solubility in mols per litre | smiles | mol | |
|---|---|---|---|---|
| 0 | Amigdalin | -0.770 | OCC3OC(OCC2OC(OC(C#N)c1ccccc1)C(O)C(O)C2O)C(O)... | <rdkit.Chem.rdchem.Mol object at 0x7b4a78f01b60> |
| 1 | Fenfuram | -3.300 | Cc1occc1C(=O)Nc2ccccc2 | <rdkit.Chem.rdchem.Mol object at 0x7b4a78f01a80> |
| 2 | citral | -2.060 | CC(C)=CCCC(C)=CC(=O) | <rdkit.Chem.rdchem.Mol object at 0x7b4a78f01af0> |
| 3 | Picene | -7.870 | c1ccc2c(c1)ccc3c2ccc4c5ccccc5ccc43 | <rdkit.Chem.rdchem.Mol object at 0x7b4a78f01bd0> |
| 4 | Thiophene | -1.330 | c1ccsc1 | <rdkit.Chem.rdchem.Mol object at 0x7b4a78f01c40> |
| ... | ... | ... | ... | ... |
| 1123 | halothane | -1.710 | FC(F)(F)C(Cl)Br | <rdkit.Chem.rdchem.Mol object at 0x7b4a78e6ca50> |
| 1124 | Oxamyl | 0.106 | CNC(=O)ON=C(SC)C(=O)N(C)C | <rdkit.Chem.rdchem.Mol object at 0x7b4a78e6cac0> |
| 1125 | Thiometon | -3.091 | CCSCCSP(=S)(OC)OC | <rdkit.Chem.rdchem.Mol object at 0x7b4a78e6cb30> |
| 1126 | 2-Methylbutane | -3.180 | CCC(C)C | <rdkit.Chem.rdchem.Mol object at 0x7b4a78e6cba0> |
| 1127 | Stirofos | -4.522 | COP(=O)(OC)OC(=CCl)c1cc(Cl)c(Cl)cc1Cl | <rdkit.Chem.rdchem.Mol object at 0x7b4a78e6cc10> |
1128 rows × 4 columns
Convert “mol” in rdkit to molecualr graphs¶
We follow this blog to convert “mol” to molecualr graphs. https://www.blopig.com/blog/2022/02/how-to-turn-a-smiles-string-into-a-molecular-graph-for-pytorch-geometric/
There are two changes compared to the implementation in the original blog:
The molecules are not embedded to 3D structures.
The edge embeddings are not considered.
[ ]:
def mol2graph(mol):
# calculate node features
ATOMS = ['C', 'O', 'S', 'N', 'P', 'H', 'F', 'Cl', 'Br', 'I', 'UNK']
ATOM2DCT = {ele: (np.eye(len(ATOMS))[idx]).tolist() for idx, ele in enumerate(ATOMS)}
HYBRIDS = [Chem.rdchem.HybridizationType.SP,
Chem.rdchem.HybridizationType.SP2,
Chem.rdchem.HybridizationType.SP3,
Chem.rdchem.HybridizationType.SP3D,
Chem.rdchem.HybridizationType.SP3D2,
'UNK']
HYBRID2DCT = {ele: (np.eye(len(HYBRIDS))[idx]).tolist() for idx, ele in enumerate(HYBRIDS)}
x = []
ring = mol.GetRingInfo()
for idx in range(mol.GetNumAtoms()):
emd = []
at = mol.GetAtomWithIdx(idx)
ele = at.GetSymbol()
ele = ele if ele in ATOMS else 'UNK'
emd += ATOM2DCT[ele] # add atom type
emd += [at.GetDegree()]
hyb = at.GetHybridization()
hyb = hyb if hyb in HYBRIDS else "UNK"
emd += HYBRID2DCT[hyb] # add atom hybridization type
emd += [at.GetIsAromatic()] # add atom aromacity
emd += [ring.IsAtomInRingOfSize(idx, 3),
ring.IsAtomInRingOfSize(idx, 4),
ring.IsAtomInRingOfSize(idx, 5),
ring.IsAtomInRingOfSize(idx, 6),
ring.IsAtomInRingOfSize(idx, 7),
ring.IsAtomInRingOfSize(idx, 8)] # add ring size
x.append(emd)
x = torch.Tensor(np.array(x)).float()
# calculate edges
bonds = []
for bond in mol.GetBonds():
idx1 = bond.GetBeginAtomIdx()
idx2 = bond.GetEndAtomIdx()
bonds.append([idx1, idx2])
bonds.append([idx2, idx1])
# create graph
edge_index = torch.Tensor(np.array(bonds)).long()
data = Data(x=x, edge_index=edge_index.t().contiguous())
return data
Visulize one of the compoudns in the dataset¶
[ ]:
mol = df.iloc[4]["mol"]
from IPython.display import SVG
def draw_single_mol(mol, size=(300, 300), **highlights):
# copy the molecule to avoid modifying the 3D coordinates
mol = Chem.Mol(mol)
drawer = Chem.Draw.rdMolDraw2D.MolDraw2DSVG(*size)
if highlights is not None:
Chem.Draw.rdMolDraw2D.PrepareAndDrawMolecule(drawer, mol, **highlights)
else:
drawer.DrawMolecule(mol)
drawer.FinishDrawing()
svg = drawer.GetDrawingText()
return svg.replace('svg:','')
def mol_with_atom_index(mol):
for atom in mol.GetAtoms():
atom.SetProp("molAtomMapNumber", f"{atom.GetIdx()}")
return mol
SVG(draw_single_mol(mol_with_atom_index(mol)))
Visulize a moleuclar graph¶
[ ]:
g = mol2graph(mol)
g
Data(x=[5, 25], edge_index=[2, 10])
Node embeddings¶
The above molecule contains 5 heavy atoms (non-hydrogen atoms). Each atom is embedded to a 25-dim vector. Therefore the node embeddings of the graph has the shape of \(5\times 25\).
[ ]:
g.x
tensor([[1., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 2., 0., 1., 0., 0., 0., 0.,
1., 0., 0., 1., 0., 0., 0.],
[1., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 2., 0., 1., 0., 0., 0., 0.,
1., 0., 0., 1., 0., 0., 0.],
[1., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 2., 0., 1., 0., 0., 0., 0.,
1., 0., 0., 1., 0., 0., 0.],
[0., 0., 1., 0., 0., 0., 0., 0., 0., 0., 0., 2., 0., 1., 0., 0., 0., 0.,
1., 0., 0., 1., 0., 0., 0.],
[1., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 2., 0., 1., 0., 0., 0., 0.,
1., 0., 0., 1., 0., 0., 0.]])
Edge list¶
The molecule has 5 bonds, but each bonds will be counted twice as \(i \to j\) and \(j \to i\) in the directed graph, resulting in 10 edges.
[ ]:
g.edge_index.T
tensor([[0, 1],
[1, 0],
[1, 2],
[2, 1],
[2, 3],
[3, 2],
[3, 4],
[4, 3],
[4, 0],
[0, 4]])
Batch compoudns for training¶
[ ]:
class MolGraphDataset(Dataset):
def __init__(self, df, mol_col="mol", target_col=TASK_COL, transform_fn=None):
super().__init__()
self.df = df
self.mol_col = mol_col
self.target_col = target_col
self.transform_fn = transform_fn
def len(self):
return len(self.df)
def get(self, idx):
row = self.df.iloc[idx]
mol = row[self.mol_col]
try:
G = self.transform_fn(mol)
if self.target_col is not None:
target = torch.tensor([row[self.target_col]], dtype=torch.float)
G.y = target
except:
return None
return G
Split Dataset¶
[ ]:
from sklearn.model_selection import train_test_split
# training/validation dataset
data_size = df.shape[0]
test_ratio = 0.10
test_size = int(data_size*test_ratio)
train_indices, test_indices = train_test_split(range(data_size), test_size=test_size, shuffle=True)
print(f"Training size: {len(train_indices)}, test size: {len(test_indices)}")
train_df, test_df = df.iloc[train_indices], df.iloc[test_indices]
Training size: 1016, test size: 112
Please note here: we are using DataLoader from torch_geometric.loader, not torch.utils.data.DataLoader. The dataloader from torch_geometric helps combine several graphs to one batch (a large graph).¶
[ ]:
# create dataloaders
batch_size = 32
train_data = MolGraphDataset(train_df, transform_fn=mol2graph)
train_loader = DataLoader(train_data, batch_size=batch_size,
shuffle=True, drop_last=False)
test_data = MolGraphDataset(test_df, transform_fn=mol2graph)
test_loader = DataLoader(test_data, batch_size=batch_size,
shuffle=False, drop_last=False)
Vislulization of graph batching (batch 5 compounds in a single graph)¶
[ ]:
# combine 5 graphs to one batch
batch = next(iter(DataLoader(train_data, batch_size=5,
shuffle=True, drop_last=False)))
print(batch)
import networkx as nx
# build a graph from the edge list of the large graph
n = batch.x.shape[0]
batch_ids = np.array(batch.batch.tolist())
edges = np.array(batch.edge_index.T.tolist())
G = nx.Graph()
G.add_nodes_from(range(n))
G.add_edges_from([edges[i] for i in range(edges.shape[0])
if edges[i][0] < edges[i][1]])
node_colors = [batch_ids[i] for i in G.nodes]
plt.figure(figsize=(5, 5))
pos = nx.spring_layout(G)
nx.draw(G, pos=pos, with_labels=False, node_color=node_colors, cmap="coolwarm",
edge_color="gray", node_size=10)
plt.show()
DataBatch(x=[63, 25], edge_index=[2, 128], y=[5], batch=[63], ptr=[6])
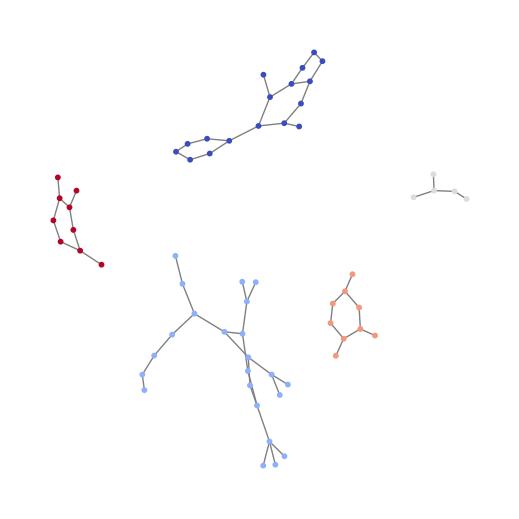
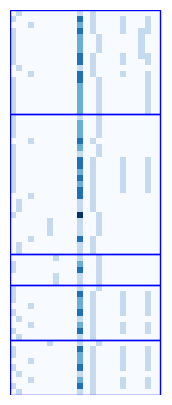
Visualize node features by batch ids¶
[ ]:
unique_batch_ids = np.unique(batch_ids)
fig, ax = plt.subplots(1, 1, figsize=(5, 5))
ax.imshow(np.array(batch.x.tolist()), cmap="Blues", interpolation='none')
for batch_id in unique_batch_ids:
indices = np.where(batch_ids == batch_id)[0]
start, end = indices[0], indices[-1]
rect = patches.Rectangle((-0.5, start-0.5), batch.x.shape[1]-0.5, end-start+1,
linewidth=1, edgecolor='blue', facecolor='none', linestyle='-')
ax.add_patch(rect)
plt.axis("off")
(np.float64(-0.5), np.float64(24.5), np.float64(62.5), np.float64(-0.5))
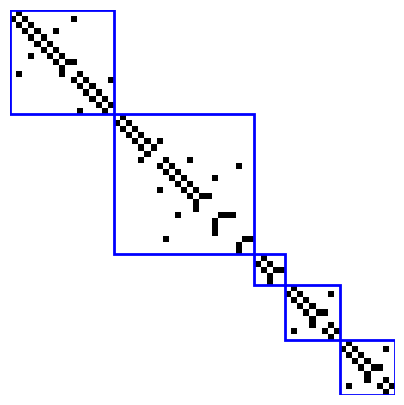
Visualize Adjacency matrix of the batched graph¶
[ ]:
def edge_list_to_adjacency_matrix(edge_list, num_nodes):
adj_matrix = np.zeros((num_nodes, num_nodes), dtype=int)
for u, v in edge_list:
adj_matrix[u][v] = 1
return adj_matrix
adj_matrix = edge_list_to_adjacency_matrix(edges, n)
print(adj_matrix)
fig, ax = plt.subplots(1, 1, figsize=(5, 5))
plt.imshow(1-adj_matrix, cmap="gray")
# add diagonal blocks
unique_batch_ids = np.unique(batch_ids)
lw = 2
for batch_id in unique_batch_ids:
indices = np.where(batch_ids == batch_id)[0]
start, end = indices[0], indices[-1]
rect = patches.Rectangle((start-0.5, start-0.5), end-start+1, end-start+1,
linewidth=lw, edgecolor='blue', facecolor='none', linestyle='-')
ax.add_patch(rect)
plt.axis("off")
[[0 1 0 ... 0 0 0]
[1 0 1 ... 0 0 0]
[0 1 0 ... 0 0 0]
...
[0 0 0 ... 0 1 0]
[0 0 0 ... 1 0 1]
[0 0 0 ... 0 1 0]]
(np.float64(-0.5), np.float64(62.5), np.float64(62.5), np.float64(-0.5))
GCN Model¶
[ ]:
class GCNModel(nn.Module):
def __init__(self, ndim, hidden_dims):
super(GCNModel, self).__init__()
total_dims = [ndim] + hidden_dims
net = []
self.bn = nn.BatchNorm1d(total_dims[0])
##set up the graph convolutional layer ndim=25 (25 node features, input channels for GCN)
#hidden-dimes: output channel of the convolutional layer
# 3 GCN layers, len(total_dims)=4
for i in range(len(total_dims)-1):
net.extend([
GCNConv(total_dims[i], total_dims[i+1], add_self_loops=True),
nn.ReLU(),
])
self.net = nn.Sequential(*net)
self.fc = nn.Linear(total_dims[-1], 1)
def forward(self, data):
batch = data.batch
out = data.x
out = self.bn(out)
edge_index = data.edge_index.long()
for idx in range(len(self.net)//2):
out = self.net[2*idx](out, edge_index)
out = self.net[2*idx+1](out)
# pool each molecule to one vector
out = global_mean_pool(out, batch)
# fully connected layer
out = self.fc(out)
return out
[ ]:
def train_one_epcoh(model, criterion, optimizer, dataloader):
losses = []
model.train()
for G in dataloader:
if device == "cuda":
G = G.to(device)
y_true = G.y
optimizer.zero_grad()
y_pred = model(G)
loss = criterion(y_pred, y_true.reshape(y_pred.shape))
loss.backward()
optimizer.step()
losses.append(loss.cpu().detach().item())
return losses
def val_one_epcoh(model, criterion, dataloader):
losses = []
model.eval()
with torch.no_grad():
for G in dataloader:
if device == "cuda":
G = G.to(device)
y_true = G.y
y_pred = model(G)
loss = criterion(y_pred, y_true.reshape(y_pred.shape))
losses.append(loss.cpu().detach().item())
return losses
Training¶
[ ]:
model = GCNModel(ndim=25, hidden_dims=[128, 64, 32]) # we stack 3 GCNs with dimensions: 128, 64 and 32
model.to(device)
model = model.float()
n_epochs = 200
lr = 5e-3 #learning rate
optimizer = torch.optim.Adam(model.parameters(), lr=lr)
print("Number of trainable parameters:",
sum(p.numel() for p in model.parameters() if p.requires_grad))
criterion = nn.MSELoss() #Mean square error loss
train_loss = []
val_loss = []
for epoch in tqdm.tqdm(range(n_epochs)):
losses = train_one_epcoh(model, criterion, optimizer, train_loader)
train_loss.append(np.mean(losses))
losses = val_one_epcoh(model, criterion, test_loader)
val_loss.append(np.mean(losses))
Number of trainable parameters: 13747
100%|██████████| 200/200 [03:10<00:00, 1.05it/s]
[ ]:
f, ax = plt.subplots(1, 1, figsize=(5,5))
ax.plot(train_loss, c="blue", label="Training")
ax.plot(val_loss, c="red", label="Test")
plt.xlabel("Epoch")
plt.ylabel("Loss")
plt.legend()
<matplotlib.legend.Legend at 0x7b4a75628e10>
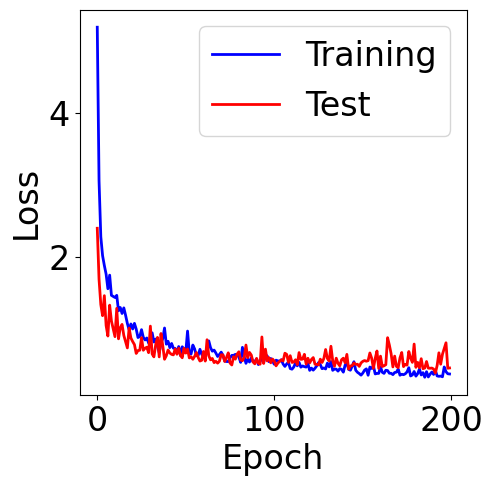
Evaluation Metrics¶
[ ]:
truths = []
predictions = []
model.eval()
with torch.no_grad():
for G in test_loader:
if device == "cuda":
G = G.to(device)
y = G.y
y_pred = model(G).reshape(-1)
# predictions.extend(y_pred.cpu().detach().numpy().tolist())
predictions.extend([y_pred[i].item() for i in range(len(y_pred))])
y = y.reshape(y_pred.shape)
# truths.extend(y.cpu().numpy().tolist())
truths.extend([y[i].item() for i in range(len(y))])
[ ]:
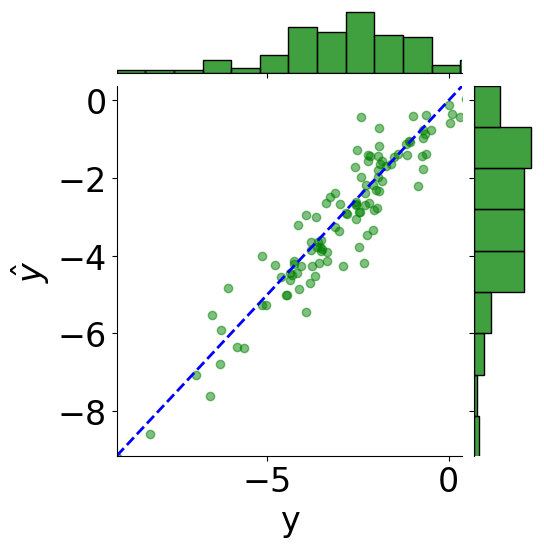
tmp_df = pd.DataFrame({"y": truths, r"$\hat{y}$": predictions})
# scatter plot
g = sns.JointGrid(x="y", y=r"$\hat{y}$", data=tmp_df)
g = g.plot_joint(plt.scatter, c="green", alpha=0.5)
# line: y_pred = y
y_line = np.linspace(np.min(truths), np.max(predictions), 200)
g.ax_joint.plot(y_line, y_line, color="blue", linestyle="--");
# histograms
g = g.plot_marginals(sns.histplot, data=df, color="green", kde=False)
g.ax_joint.set_xlim(np.min(y_line), np.max(y_line))
g.ax_joint.set_ylim(np.min(y_line), np.max(y_line))
plt.show()
[ ]:
from sklearn.metrics import r2_score
from sklearn.metrics import mean_squared_error
print(f"MSE: {mean_squared_error(truths, predictions):.2f}")
print(f"Coefficient of determination: {r2_score(truths, predictions):.2f}")
MSE: 0.43
Coefficient of determination: 0.88
[ ]: